ALAS! Why is gov’t data so terribly structured?!
I’ve been working with the CMS Open Payments data recently because the annual datasets offer enough granular info to allow for good modeling practices and pharmaceutical marketing practices are pretty interesting from a policy stand-point.
My only harp with the data is it’s poor data structure and lack of standardization. Aside from running DQA and standardization scripts, I’ve been fairly involved in subdividing the original dataset into relational data tables that are much easier to work with. One of the most involved tasks I had to do was write conditional reformatting scripts for payment detail scripts because CMS changed the release data-structure between 2015 & 2016.
It’s not like the data structure was pretty begin with, and although the new structure is a little easier to work with than what was used in PY13-15, the abrupt switch definitely adds a few extra headaches in preparing the data for analysis.
Here’s what the old structure looked like:

Here is the new one:

The drawback of the legacy structure was that it lumped the Medicare/Medicaid coverage information into a single “Product Indicator” field when up to 10 products could be reported for a single transaction, and reports their joint coverage as ‘Covered’, ‘Non-covered’ or ‘Mixed’.
The new 2016 structure fixed this issue by splitting coverage information into independent ‘Covered_or_Noncovered_Indicator’ fields, but the encapsulation of the one-to-many relationship between transactional and product information in a single row still made running queries inefficient.
To get over this hurdle, I had to write separate conditional sorting protocols – one for PY2013-2015 and one for 2016 due to varying levels of data granularity.
Here’s a quick mock up of the sorting protocol for 2013-2015:
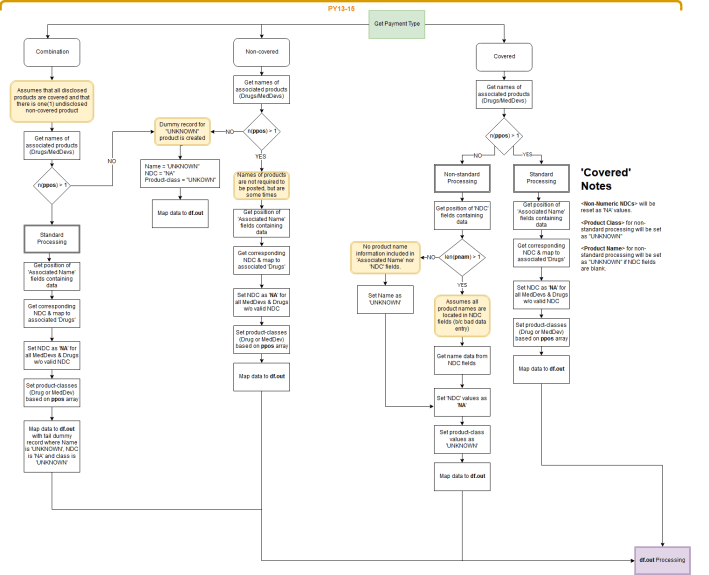
Yeah, man … not pretty.
The most efficient method I found to write & test the procedure was individually process each transactional record into its own small dataframe and store it into a pre-allocated list.
That way, I could easily do spot checks between original transaction records and their corresponding reformatted dataframes by referencing the same object number in the CMS dataset and output list. Although this method is extremely fast if you wrap your sorting logic into a custom function and then process everything using an apply function, it has some serious drawbacks.
Specifically, you get massive list object in the end.
Here's a sample of the code...
# Create temporary dataframe to hold non-product transactions
pydet_13.npp = as.data.frame(matrix(NA, nrow = nrow(pydat_13[pydat_13$Product_Indicator == "None",]), ncol = 6))
names(pydet_13.npp) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
pydet_13.npp$Payment_ID = pydat_13$Payment_ID[pydat_13$Product_Indicator == "None"]
pydet_13.npp$Payment_Type = "NONPRODUCT"
# Create temporary data-frame list to hold product transactions
ppp = function(payment){
if(payment$Product_Indicator == "Covered"){
# Sort Logic for <> payment txns
# ... 1) Determine the number of mentioned products
# ... if >1 then continue with normal processing
# ... if <1 then assume that name contents are located within the NDC-fields
# Determine the Number of Mentioned Products
ppos = which(is.blank(payment[1,grep("^Name_of_Associated", names(payment))]) == FALSE)
# :: Relative Table Position of Product Names
# 1-5 = Drug/Biological
# 6-10 = Medical Device/Supply
if(length(ppos) >= 1){
# Get Product Names
pnam = as.character(payment[1,grep("^Name_of_Associated", names(payment))][ppos])
# Get Corresponding NDC Data
pndc = as.character(payment[1, grep("^NDC_of_Associated", names(payment))][ppos[ppos < 6]]) pndc = c(pndc, rep(NA, (length(pnam) - length(pndc)))) # Match array length to 'pnam' # Validate NDC Values; should contain only numeric characters pndc[grep("[^[:digit:]]",pndc)] = NA pndc[pndc == ""] = NA # Set Corresponding Product Classes pcls = rep("Drug/Biological", length(ppos)) pcls[which(ppos > 5)] = "Medical Device/Supply"
# Create Output Dataframe
df.out = as.data.frame(matrix(NA, nrow = length(ppos), ncol = 6))
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class = pcls
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name = pnam
df.out$Product_NDC = pndc
}else {
# Condition where names are inappropriately included in NDC-field
# Quantified as less than .01% of records in PY13
# Get names in NDC Field
ppos = which(is.blank(payment[1,grep("NDC_of_Associated", names(payment))]) == FALSE)
pnam = as.character(payment[1,grep("NDC_of_Associated", names(payment))][ppos])
if(length(pnam)<1){pnam = "UNKNOWN"; ppos = 1} # When no name information is available
# Set Class as 'Unknown' because both drugs & devices have been observed in error fields
pcls = rep("UNKNOWN", length(pnam))
# Create Output Dataframe
df.out = as.data.frame(matrix(NA, nrow = 1, ncol = 6))
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class = "UNKNOWN"
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name = pnam
df.out$Product_NDC = rep(NA, length(ppos))
}
}else if(payment$Product_Indicator == "Non-Covered"){
# Determine the Number of Disclosed Products
# ...If products are disclosed products, they assumed to be 'Non-Covered' products
# ...If no products are disclosed, a dummy entry is made for one (1) 'Undisclosed' product,
# though payment maybe associated with more than one. Unable to determine this fact.
ppos = which(is.blank(payment[1,grep("^Name_of_Associated", names(payment))]) == FALSE)
# :: Relative Table Position of Product Names
# 1-5 = Drug/Biological
# 6-10 = Medical Device/Supply
if(length(ppos) >= 1){
# Condition: Product names are disclosed
# Use standard 'covered' product sorting method, with necessary adjustments
pnam = as.character(payment[1,grep("^Name_of_Associated", names(payment))][ppos])
# Get Corresponding NDC Data
pndc = as.character(payment[1, grep("^NDC_of_Associated", names(payment))][ppos[ppos < 6]]) pndc = c(pndc, rep(NA, (length(pnam) - length(pndc)))) # Match array length to 'pnam' # Validate NDC Values; should contain only numeric characters pndc[grep("[^[:digit:]]",pndc)] = NA pndc[pndc == ""] = NA # Set Corresponding Product Classes pcls = rep("Drug/Biological", length(ppos)) pcls[which(ppos > 5)] = "Medical Device/Supply"
# Create Output Dataframe
df.out = as.data.frame(matrix(NA, nrow = length(ppos), ncol = 6))
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class = pcls
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name = pnam
df.out$Product_NDC = pndc
}else{
# Condition: Product names are not disclosed
# Create dummy-record for 'Undisclosed' product
df.out = as.data.frame(matrix(NA, nrow = 1, ncol = 6))
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class = "UNKNOWN"
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name = "UNKNOWN"
df.out$Product_NDC = NA
}
}else if(payment$Product_Indicator == "Combination"){
# Assumes that all disclosed products are covered and that there is one(1) undisclosed non-covered product
ppos = which(is.blank(payment[1,grep("^Name_of_Associated", names(payment))]) == FALSE)
# :: Relative Table Position of Product Names
# 1-5 = Drug/Biological
# 6-10 = Medical Device/Supply
if(length(ppos) >= 1){
# Condition: Product names are disclosed
# Use standard 'covered' product sorting method, with necessary adjustments
pnam = as.character(payment[1,grep("^Name_of_Associated", names(payment))][ppos])
# Get Corresponding NDC Data
pndc = as.character(payment[1, grep("^NDC_of_Associated", names(payment))][ppos[ppos < 6]]) pndc = c(pndc, rep(NA, (length(pnam) - length(pndc)))) # Match array length to 'pnam' # Validate NDC Values; should contain only numeric characters pndc[grep("[^[:digit:]]",pndc)] = NA pndc[pndc == ""] = NA # Set Corresponding Product Classes pcls = rep("Drug/Biological", length(ppos)) pcls[which(ppos > 5)] = "Medical Device/Supply"
# Create Output Dataframe
df.out = as.data.frame(matrix(NA, nrow = (length(ppos) + 1), ncol = 6))
df.len = nrow(df.out)
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class[1:(df.len - 1)] = pcls
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name[1:(df.len - 1)] = pnam
df.out$Product_NDC[1:(df.len - 1)] = pndc
df.out$Product_Class[df.len] = "UNKNOWN"
df.out$Product_Name[df.len] = "UNKNOWN"
df.out$Product_NDC[df.len] = NA
}else{
# Condition: Product names are not disclosed
# Create dummy-record for 'Undisclosed' product
df.out = as.data.frame(matrix(NA, nrow = 1, ncol = 6))
names(df.out) = c("Payment_ID","Payment_Type","Product_Class","Product_Coverage", "Product_Name", "Product_NDC")
df.out$Payment_ID = payment$Payment_ID[1]
df.out$Payment_Type = "PRODUCT"
df.out$Product_Class = "UNKNOWN"
df.out$Product_Coverage = payment$Product_Indicator[1]
df.out$Product_Name = "UNKNOWN"
df.out$Product_NDC = NA
}
}else{
#Return Warning for Unknown payment-type
warning(paste("Payment_ID:", as.character(payment$Payment_ID[1]), "", sep = " "))
}
df.out
}
# Payment Processsing Test -> PYDAT_13 txns
prodlist = which(pydat_13$Product_Indicator != "None"); #rm(a,b)
paylist_13 = vector(mode = "list",length = length(prodlist))
system.time({
for(i in 1:length(prodlist)){
paylist_13[[i]] = ppp(pydat_13[prodlist[i], ])
if((i%%10000) == 0){print(i)}
}
})
The 2013 data contained approximately 3M transactions, resulting in an 8.4 Gb output list of data-frames.
Besides the inherent query challenges associated with lists of dataframes, they are a pretty impractical way to store information.
To resolve this, I had to reprocess this list into a single dataframe object, but the traditional methods of do.call("rbind", listofdataframes) and ldply(listofdataframes, data.frame) were too slow, sharply decreasing in efficiency for objects greater than 500 Mb.
I tried them initially and then gave up after sitting and staring at processing screen for 20 minutes.
Luckily, there’s a much faster method: as.data.frame(data.table::rbindlist(listOfDataFrames))
The ‘new’ data.table package method created the final dataframe in about 5 minutes resulting, and the dataframe was only 0.4 Gb in size.
That’s a 95% reduction in object size. — I wasn’t expecting that, but it makes sense conceptually since each individaul dataframe has a fixed memory cost that adds up eventually as you create hundreds of thousands of them.
I bet that if I had let the do.call method run it’s course it would have taken at least 1.5hrs.
If you’re interested in actual comparative benchmarks between the various methods, here’s a snippet from the stackoverflow page where I found this technique:
library(rbenchmark)
benchmark(
do.call = do.call("rbind", listOfDataFrames),
plyr_rbind.fill = plyr::rbind.fill(listOfDataFrames),
plyr_ldply = plyr::ldply(listOfDataFrames, data.frame),
data.table_rbindlist = as.data.frame(data.table::rbindlist(listOfDataFrames)),
replications = 100, order = "relative",
columns=c('test','replications', 'elapsed','relative')
) .... test replications elapsed relative
4 data.table_rbindlist 100 0.11 1.000
1 do.call 100 9.39 85.364
2 plyr_rbind.fill 100 12.08 109.818
3 plyr_ldply 100 15.14 137.636Here’s a glimpse of what the final output datatable looks like:
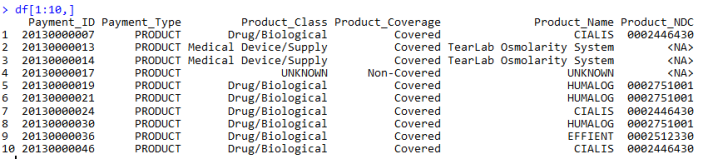
All in all, I am happy with the results. It’s safe to say I never want to go through this ordeal ever again (definitely will).
[…] noted in a previous post that the CMS data-structure is horrendous, so it was further processed to enable efficient […]
LikeLike